Single Cell Genomics
Services
Our facility provides services in single-cell and spatial omics technologies to BRIC and non-BRIC users:
- Droplet-based 10x Genomics platforms
- Split-pool barcoding using Parse Biosciences platforms
- Smart-seq2
- 10x Genomics Visium Spatial Transcriptomics
Users are responsible for nuclei/cell isolation and fixation, depending on the chosen technology (see subpage TECHNOLOGIES).
The service consists of the following blocks that can be ordered together or separately:
- Preparation of libraries from provided cell/nuclei suspension or tissue sections (Visium)
- Sequencing of prepared libraries
- Single-cell computational analysis
Don’t hesitate to contact us if you have any questions.
Our equipment
- 10X Genomics Chromium X
- Sciclone G3 NGSx iQ™ Workstation (PerkinElmer)
- PCR Workstations with HEPA Filtration and UV
- Microscope for Visium technology
- MANTIS® automated liquid handler
- Curiox Laminar Wash MINI1000 System
We also have access to
- 10x CytAssist
- Illumina NextSeq550 and NovaSeq6000 sequencing systems
Droplet technology (10x Genomics)
 Combinatorial barcoding (Parse)
Combinatorial barcoding (Parse)
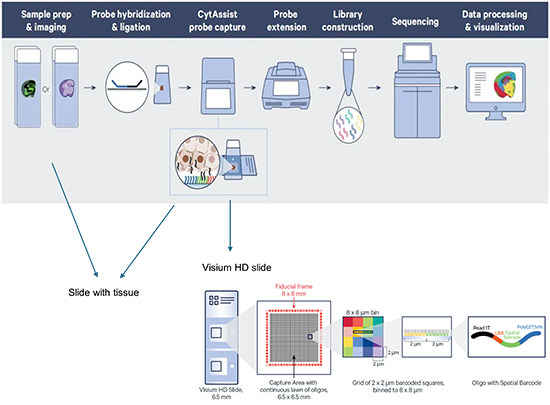
 Visium HD technology
Visium HD technology
|
Technology |
10x Genomics Universal 3’ Gene Expression |
10x Genomics Flex Gene Expression |
PARSE Biosciences |
Smart-Seq 2 and Patch-Seq* |
10x Genomics Epi Multiome ATAC + Gene Expression |
Spacial transcriptomic Visium HD WT (probe-based) |
|---|---|---|---|---|---|---|
|
Input material |
Cell/nuclei suspension Freshly prepared from tissue (no fixation) |
Cell/nuclei suspension Fixed and kept at -80 °C |
Cell/nuclei suspension Fixed and kept at -80 °C |
Cell/nuclei suspension Fresh frozen |
Cell/nuclei suspension Freshly prepared from tissue |
Slide with tissue
|
|
Short description |
1Droplet technology Whole transcriptome profiling Transcripts are captured by poly(dT) oligos on 10x gel beads and receive a cell-specific barcode and UMI during reverse transcription |
1Droplet technology Probe-based single-cell transcriptome profiling for fixed samples. Transcripts are captured and barcoded by a probe set, which covers only endogenous genes. Exogenous genes require custom probes, and only ligated probes are sequenced. |
2Combinatorial barcoding Whole transcriptome profiling Transcripts are captured by poly(dT) oligos Split-pool combinatorial barcoding of fixed samples |
One cell – one well Captures the full-length mRNA full-length capture is needed for studies concerned with isoforms or gene fusions |
1Droplet technology Captures the transcriptome and chromatin accessibility profiles Direct linkage of gene expression and open chromatin readouts from the same nucleus Aim to discover new gene regulatory interactions |
3Microscope + CytAssist Assigning cell types (identified by the mRNA readouts) to their locations in the histological sections and can also be used to determine subcellular localization of mRNA molecules |
|
Kind of tissue |
Fresh or frozen tissue |
Fresh, frozen, FFPE, PFA-fixed tissue |
Fresh or frozen tissue |
Fresh or frozen tissue |
Fresh or frozen tissue |
Fresh frozen, FFPE, Fixed frozen |
|
Species restraints |
none (currently shown on mouse, human, rat, primates, marine, plant, insect) |
mouse and human (other species with custom probes) |
none |
none |
mouse, human, rat, primates, marine, plant, insect |
mouse and human |
|
Sample number limitations |
1-8 samples per run
Up to 10000 cells per sample |
4 - 128 samples per run 4-16 samples can be multiplexed in each reaction. Up to 20,000 cells/nuclei can be targeted per probe barcode (sample) |
1- 96 samples per run |
1-24 samples per run (potentially up to 96) |
1-8 samples per run |
2 samples per run |
|
Final concentration of fresh or fixed suspension |
The fresh nuclei suspension needs at least a concentration of 1,000-2,000 nuclei/µl in a volume of at least 80µl. Max Load: 16,000 cells/nuclei (with aim to recover 10,000 nuclei)
|
Fixation protocol – get 1 x 10^6 cells For proceeding with probe hybridisation – deliver to us ca 300,000 cells distributed between 2 aliquots |
Fixation protocol – get 1 x 10^6 cells |
1 cell per well |
The fresh nuclei suspension needs at least a concentration of 4,000-8,000 nuclei/µl Max Load: 10,000 nuclei (with aim to recover 7,500 nuclei)
|
Specific requirements for Fresh frozen, FFPE, Fixed frozen tissue slides |
|
Key advantage |
Standard, widely used Greater consistency in comparison to Parse |
Fixed samples, flexible handling, nuclei can be obtained from frozen, FFPE and PFA- tissue Greater consistency in comparison to Parse |
Less expensive, large-scale studies, fixed samples, flexible handling Can detect long non-coding RNA Almost no ambient RNA (washed away during prep) Does not need a special device |
Unique technology |
Unique technology |
Unique technology |
|
Disadvantage |
Limited numbers of samples can be processed per day Expensive |
Only compatible with mouse and human samples (but can work with custom made probe) |
For sequencing – more expensive (more reads/cell) Lower cell recovery rate Lower sensitivity for genes with high GC content |
Expensive, much fewer cells are processed |
Expensive |
Expensive |
*Patch-Seq
Patch-seq experiments profile the electrophysiological properties and transcriptome of the same individual neurons, with the goal of identifying the underlying relationships between gene expression and neuronal function.
Additionally, recorded neurons can be backfilled with appropriate dies to evaluate cell morphology.
You
- perform electrophysiology part
- aspirate neuronal content and snap freeze
- send to our facility
We
- provide you protocol including special conditions of electrophysiology part, neuronal collection, and shipment
- perform RNA Isolation, cDNA Synthesis, and Library Preparation
- sequence cDNA libraries
Links to more detailed description of technologies
Single cell omics
- 10x Genomics Universal 3’ Gene Expression (no sample fixation)
- 10x Genomics Universal 5’ Gene Expression (no sample fixation)
- 10x Genomics Flex Gene Expression with sample fixation and multiplexing of samples (only human or mouse samples)
- 10x Genomics Epi Multiome ATAC + Gene Expression
- Parse EvercodeTM WT Mini (up to 12 samples), EvercodeTM WT (up to 48 samples), and EvercodeTM WT Mega (up to 96 samples) with sample fixation
- Smart-seq2 (full-length transcript coverage) : example
- Patch-seq: original paper 1 and paper 2
Spatial omics
In single-cell transcriptomics, careful experimental design is crucial to minimize batch effects and confounding factors that could lead to spurious clustering and mask the true biological signal. Ensuring an adequate number of samples and incorporating appropriate controls tailored to your research question is equally important to generate robust and interpretable data.
This guide provides key considerations for designing your single-cell experiment. We do not take responsibility for the completeness of the information provided. Always adapt your experimental design to the specifics of your study.
What is a batch effect?
A batch effect refers to systematic differences in your data caused by technical factors rather than biological variation. These effects can arise due to:
- Differences in sample preparation protocols
- Optimize your sample preparation protocol and maintain consistency! Always use the same reagents and equipment.
- The person performing the sample preparation
- Ideally, a single person should handle all samples to minimize variability.
- The day of sample preparation
- Process as many samples as possible per day to reduce variability across batches.
Some batch effects are unavoidable, such as those caused by processing samples on different days or involving multiple individuals in sample preparation. To mitigate their impact, it is essential to:
- Record all batch-related variables (e.g. date, operator, reagent lot numbers).
- Ensure a balanced experimental design, distributing conditions (e.g. treatment, genotype) and biological covariates (e.g. sex, age, post-mortem interval) evenly across batches.
- Address batch effects during data analysis if necessary.
How to design your single-cell experiment
- Define sample groups and controls
- Determine the number of samples per group based on your research question.
- Ensure appropriate controls (e.g., age-matched samples).
- Identify key biological covariates
- Common covariates include sex, age, treatment, genotype, and post-mortem interval (for human samples).
- Plan sample preparation to minimize batch effects
- Distribute covariates evenly across batches.
- Avoid processing only control samples on one day or all younger samples first.
- Create a sample processing schedule and document it for downstream analysis.
By carefully planning and recording your experiment, you can reduce batch effects and improve the reliability of your single-cell data.
Proper preparation of cells/nuclei from tissue is a crucial step for all single-cell methods. We are not able to perform this step on your behalf, as different tissue and cell types require different isolation protocols. Users are expected to provide a cell/nuclei suspension, which we will process further for library preparation.
To ensure optimal results, the submitted cell/nuclei suspension must meet the following criteria:
- ≥70% viable cells (however, viability is not relevant in case of nuclei preparation when you have frozen or fixated tissue)
- Free of visible debris and aggregates
Excess debris or clumped cells can negatively affect data quality and may clog the microfluidic chip during cell loading (10x Genomics platform), which can prevent successful cell recovery and result in experiment failure.
Please consult our recommended protocols on the Resources subpage.
Cell sizes limitation for 10x Genomics droplet technology
Cells can have a diameter of up to 30 µm. Cells larger than 30 µm increase the risk of chip clogging and should not be processed using 10x droplet-based methods. For samples containing large cells, Parse Biosciences workflow, which has no cell size restrictions, is recommended.
FACS and 10x Genomics droplet technology (without fixation)
FACS-sorted samples are compatible with 10x Genomics workflow. FACS is an effective method to enrich for specific cell populations based on cell surface markers, and to remove dead cells and debris, resulting in a cleaner cell suspension.
Note: Cell counts obtained by FACS are often overestimated. We strongly recommend re-counting cells after sorting f.e. using a microscope.
We offer computational analysis and consultation for single cell data from multiple platforms, such as 10x and Parse Biosciences.
We strongly recommend contacting us as early as possible in your experimental design.
Below, in the section “Standard analysis”, there is a suggestion of the most common steps in the analysis of single cell data and the expected number of hours in a simple type of dataset which would involve one condition or two conditions to be compared. For more complex datasets, please contact us and we can make a better estimate for your particular case.
Further types of analyses can be done on single cell data depending on your needs. In section “Further analyses” you can find suggestions of analyses that we could help you with. Please, don’t hesitate to contact us if you are interested in something else as we may not include all the possibilities in this list or to discuss the estimation of number of hours needed for each analysis. Also note that the time dedicated to meetings or communications is not included and will need to be taken into account.
Standard analysis
Raw data processing [5 hrs]
- Set up the computational environment
- Produce count matrices
- Initial quality control of sequencing and cell calling
Quality control of cells
- Remove low-quality cells, typically based on UMI counts and mitochondrial read fraction [3 hrs]
- Remove potential doublets [3 hrs]
Initial data analysis
- Normalization, pre-processing, integration of different samples/conditions and dimensionality reduction [3 hr]
- Overview of sample differences [1 hr]
Cells clustering and annotation
* Note: this part of the analysis can be done multiple times until the user is satisfied with the results. The number of hours shown for each step is the expected maximum for all the expected iterations. If less hours are required, the user will be billed just for the hours that have been used. Alternatively, if the number of hours is going to be surpassed for the analysis the user will be informed beforehand.
- Cells clustering [5 hr]
- Cell type annotation in collaboration with the user [20 hrs]
Cell type specific differential analysis
* Note 1: this part of the analysis can be done multiple times as a continuation of cell cluster and annotation. The number of expected hours indicated in this section is only for 1 iteration.
* Note 2: The number of expected hours indicated in this section is only for 1 comparison between 2 conditions.
- Transcriptomic expression shifts [1 hr]
- Differentially expressed gene analysis [1 hr]
- Gene set enrichment analysis [2.5 hr]
Further analyses
- Gene network analysis (with WGCNA)
* Note 1: this part of the analysis can be done multiple times as a continuation of cell cluster and annotation. The number of expected hours indicated in this section is only for 1 iteration by the user.
* Note 2: The identification of gene modules may involve an iterative process by the bioinformatician in which different parameters will be tested to obtain the best results. The number of hours indicated shows the maximum expected number of hours for the first iteration. Further iterations after discussions with the user may be shorter. Variations on this number will be notified to the user in due time.
- Identification of gene modules [20 hr]
- Cluster-free differential analysis (using cacoa)
- Cell-cell interactions (using CellChat)
- Cellular trajectory analysis (using scVelo)
Prices
BRIC users: 400 DKK/hour
UCPH users: 500 DKK/hour
External users: 980 DKK/hour
Project data storage fee: 82 DKK per Tb per month.
BRIC autonomous users
Or contact us by email: Irina.korshunova@bric.ku.dk
Please schedule any single cell experiment at least 2 weeks in advance.
If you have no experience, consultation is necessary before planning an experiment.
Info for non-BRIC users
Contact us by email to discuss your project Irina.korshunova@bric.ku.dk
We will also need this info from you:
- Your name
- Your email
- Affiliation
- Adress
- Leader group name
- Leader group email
- CVR/VAT no.
- EAN
- Optional: Contact information for the accountant-manager
Prices for BRIC-users
Last update: January 2026
The provided here examples are for informational purposes. The exact cost is calculated individually for each specific experiment.
|
10x Universal 3’ Gene Expression v3.1 Next GEM |
4 samples |
8 samples |
16 samples |
|---|---|---|---|
|
Reagents |
56,000 |
112,000 |
224,000 |
|
Sequencing 20,000 reads/cell x 10,000 cells (kit+machine time) |
23,400 |
39,600 |
69,300 |
|
Our job approximately |
6,000 |
12,000 |
24,000 |
|
Summary |
85,400 |
163,600 |
317,300 |
|
10x ATAC multiome |
4 samples |
8 samples |
16 samples |
|---|---|---|---|
|
Reagents |
99,000 |
198,000 |
350,000 |
|
Sequencing ATAC 25,000 reads/cell x 10,000 cells (kit+machine time) |
39,600 |
69,300 |
69,300 |
|
Sequencing gene expression 20,000 reads/cell x 10,000 cells (kit+machine time) |
23,400 |
39,600 |
69,300 |
|
Our job approximately |
8,000 |
16,000 |
32,000 |
|
Summary |
170,000 |
322,900 |
520,600 |
|
Parse Evercode |
WT Mini kit 10,000-20,000 cells or nuclei per kit (up to 12 samples) |
WT Mdi kit 100,000 cells or nuclei per kit (up to 48 samples) |
WT Mega kit 1,000,000 cells or nuclei per kit (up to 96 samples) |
|---|---|---|---|
|
Reagents for wetlab protocol (*without fixation) |
35,300 |
83,500 |
141,500 |
|
Sequencing 30,000 reads/cell (kit+machine time) |
23,400 |
73,000 |
342,000 |
|
Our job approximately |
5,000 |
8,000 |
16,000 |
|
Summary |
63,700 |
164,500 |
499,500 |
*Price of reagents for fixation of cells or nuclei depends on amount of samples to be loaded on Parse kit, it should be estimated additionally.
Fixation of 1 sample - 300 DKK
|
10x Genomics FLEX |
4 samples |
16 samples |
64 samples |
|---|---|---|---|
|
At! Kits are available only for human or mouse cells |
43,000 |
114,700 |
275,000 |
|
Sequencing 10,000 reads/cell x 10,000 cells (kit+machine time) |
23,400 |
39,600 |
114,000 |
|
Our job approximately. |
6,000 |
18,000 |
36,000 |
|
Summary |
72,400 |
172,300 |
425,000 |
|
10x Visium HD (probe-based) |
2 samples |
4 samples |
8 samples |
|---|---|---|---|
|
At! Kits are available only for human or mouse cells |
48,000 |
96,000 |
192,000 |
|
Sequencing costs are variable. Depends on capture area. For full capture area: |
23,400 |
39,600 |
69,300 |
|
Our job approximately (including CytAssist run) |
9,000 |
18,000 |
36,000 |
|
Summary |
80,400 |
153,600 |
297,300 |
|
Smart Seq |
Example for 110 cells |
||
|---|---|---|---|
|
Wet lab protocol - Reagents and our Job |
78,000 |
||
|
Sequencing |
13,527 |
||
At! Note that Single Cell Genomics facility (SCG) cannot cover the cost in the case of failure. If the failure is due to manufacturing flaws of the 10X components or the Illumina system, the companies will compensate.
Protocols for nuclei/cell fixation are available on the company's webpages
- Cell Preparation Guide – general recommendations
- 10x Genomics FLEX
- Parse Biosciences:
Our in-house protocols for nuclei preparation
- Nuclei isolation with gradient
Other helpful links
- 10x Genomics support – protocols, Q&A, video etc
- Parse user guides
- Illumina Flowcell Capacity Calculator
- Illumina NovaSeq 6000 Sequencing kits
Contact to Parse and 10x Genomics representatives in Denmark
Parse
Sales Manager:
Anne Helness, anne@parsebiosciences.com
Field application scientist:
Michael Palmer, mpalmer@parsebiosciences.com
10x Genomics
Sales Manager:
John Wahlgren, john.wahlgren@10xgenomics.com
Field application scientist:
Aydan Derdiyok, aydan.derdiyok@10xgenomics.com
Please recognize and acknowledge Single Cell Genomics facility (SCG) contributions to the scientific improvements in your project. Being acknowledged in publications is very important as it serves as documentation for SCG value and performance.
Remember to notify us when a SCG acknowledgement is published.
Acknowledgment section
“We acknowledge [X], [Y], [Z] and the Single Cell Genomics facility for technical and computational expertise and support.” (Please [insert] the names of SCG members that have helped you.)
Material and Method section
Contact SCG if you need a method description of the provided NGS service(s) and please add the following acknowledgment:
“_____________ was performed by Single Cell Genomics facility (BRIC, University of Copenhagen)”
Contact
Irina Korshunova
Irina.korshunova@bric.ku.dk
BRIC - Biotech Research & Innovation Centre
Ole Maaløes Vej 5
DK-2200 Copenhagen
Room 3-4-18 and Room 3.4.20 or in Khodosevich group office space (building 3, 4th floor).

Associate Professor, Group leader,
konstantin.khodosevich@bric.ku.dk

PhD, Staff scientist,
Irina.korshunova@bric.ku.dk

Technician,
benteem@sund.ku.dk
